Give LLMs memory with RAG
-
 Nicholas Charriere
Nicholas Charriere
- Date
LLMs are goldfish
Despite their magic, we’ve all brushed up against the limits of LLMs. You probably have September 2021 burned into your mind now, as I do, as the date history stopped, at least as far as ChatGPT goes. Similarly, LLMs don’t know about things they can’t know about - like your company’s internal documentation, or your private notes. There is a way around this, and it’s also the technique used for all of those “chat with your Data” or “chat with your PDF” applications.
It’s called Retrieval Augmented Generation (RAG for short), and it’s conceptually quite simple and clever. Let’s dig into how it works.
Let’s work off a use case where we want the AI to answer questions about something that it would usually not do too well at:
Alright, first let’s take the birds eye view: what does a RAG workflow look like?
The idea is that we first ingest documents into a database, and then retrieve the relevant ones when a user asks a question. Finally, we add the relevant information to the prompt, and get a much better answer.
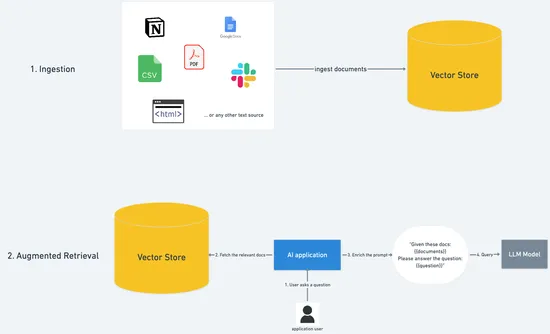
Let’s dive into the specifics of the ingestion phase first, and then we’ll look at the augmented retrieval.
Ingestion
The goal here is to get text data from a data source (this could be anything: PDFs, CSV, HTML web pages, the constitution, etc…), and prepare it for the later stages.
A first issue is that the data size is potentially unbounded: how big is a pdf or a google doc? Ingesting the whole Lord of the Rings book would be too much to handle for a single row in most databases.
The solution to the arbitrary size of the text documents is chunking. We split the text document into smaller parts, which are bounded in size. There are many chunking strategies, which depend heavily on the structure of the document being ingested:
- You might chunk slack messages by putting one message per chunk
- You might chose a fixed number of words (or tokens) and chunk that way
- You might chunk a textbook by chapters and sections
- You might want to chunk a CSV by splitting each row into its own chunk…
It’s worth noting here that chunking libraries are quite non-trivial, and you might want to spend extra time on this if you’re building such a workflow. How you would extract the info from a GitHub issue is fundamentally different to how you would chunk a wikipedia article, or a notion document.
Alright, so now that we have chunks, how do we store them for later retrieval?
Let’s remember that our goal is to answer questions with as much relevance as possible, for instance if a user asks about the weather in San Francisco, we need to be able to find the chunks that have information about… the weather in San Francisco!
For this we use a similarity search algorithm, over embeddings. I won’t go too far into what embeddings are, but briefly: they are a vector representation (a one dimensional array of floats) of the “essence” of the text chunk. They capture meaning rather than letters, so “fruit” and “apple” are close in their embedding space, but “car” and “apple” are not.
We achieve this by extracting a middle layer from a neural network. If you want to learn more, this article is a great overview.
So we take a chunk of text, and get the embedding vector for it:
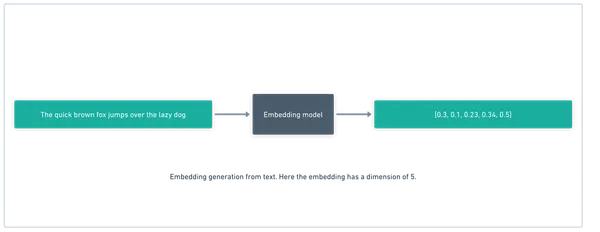
Great, now we have fixed size vectors that are very well suited for storage. We store them in a vector DB (a database optimized for this type of data shape like pinecone, chroma, or postgres with the pgvector extension).
There it is, we have successfully performed the ingestion step, let’s move on to retrieval.
Retrieval
Let’s go back to our example. Say we’ve ingested the wikipedia page about San Francisco. We have split it into chunks, transformed those into vector embeddings, and stored them in a vector DB.
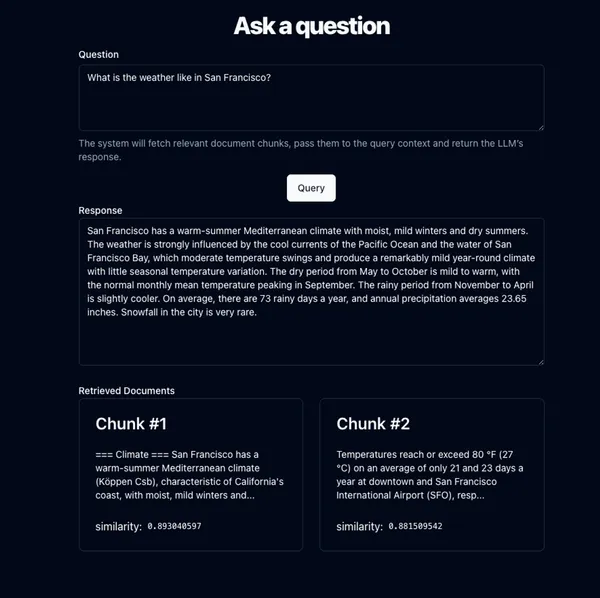
Now when a user asks a question about San Francisco, like “What is the weather like in San Francisco?”, we want to find the relevant chunks, so we can pass them to our prompt.
For this we use a similarity search algorithm over the embeddings vector space, since we want to find pieces of text that share similar meaning to the query. There are lots of good algorithms for this (FAISS, Annoy are two popular choices created by Meta and Spotify respectively).
We take the user’s query, get the embeddings representation for it in the exact same way we did for the chunks during ingestion, and finally run the similarity search algorithm. This gives us a ranked list of the top K most relevant chunks (we can configure how large K is, trading off latency, size, and accuracy).
We’re getting closer! Now we need to add the chunk text to the LLM prompt, and query the completion AI inference endpoint (for example by calling chatGPT API). You could write many kinds of prompt, but they would look something like this:
Given the following context, please answer the question below:
Context:{{chunk data: The weather in San Francisco…}}
Question:
{{user question: What’s the weather like in San Francisco?}}Your Answer:
And now, the LLM typically does a great job of answering the question, since it has all of the relevant information right there in the prompt for it to give an augmented answer based on the retrieved context. 🎉
Conclusion
RAG workflows are conceptually simple, and a very powerful way to augment LLMs with the knowledge they need to answer domain specific questions.
We built a demo of this, and it’s open source. You can find a video of me showing the San Francisco example here (1min).
Building all of this from scratch is doable, but a lot of work. This is where our axgen module comes in! With axgen, we give you different data loaders, chunking strategies, model integrations, embedding models, vector stores integrations, etc…
npm install axgenWith our library, you can build a RAG workflow in TypeScript very easily, and focus on the product, not the boilerplate:
- You could build a chrome extension that ingests the current webpage and can answer any question about it or summarize it!
- You could build a web app that ingests all of your personal notes and can help you find your thoughts and essays!
We’re excited to see what you build with it, so please reach out with any questions or cool products, and give axgen a star on GitHub if you like it, it helps us a lot.